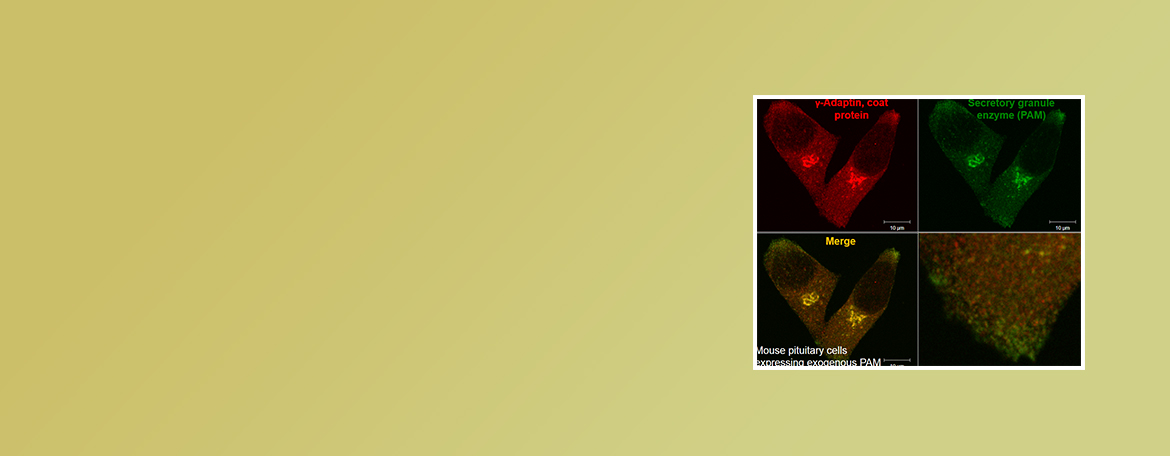
Eipper Lab
Neuropeptides are used to control many functions including mood, appetite and water balance. We are interested in dietary and genetic factors that affect the ability of cells to produce these signaling molecules.
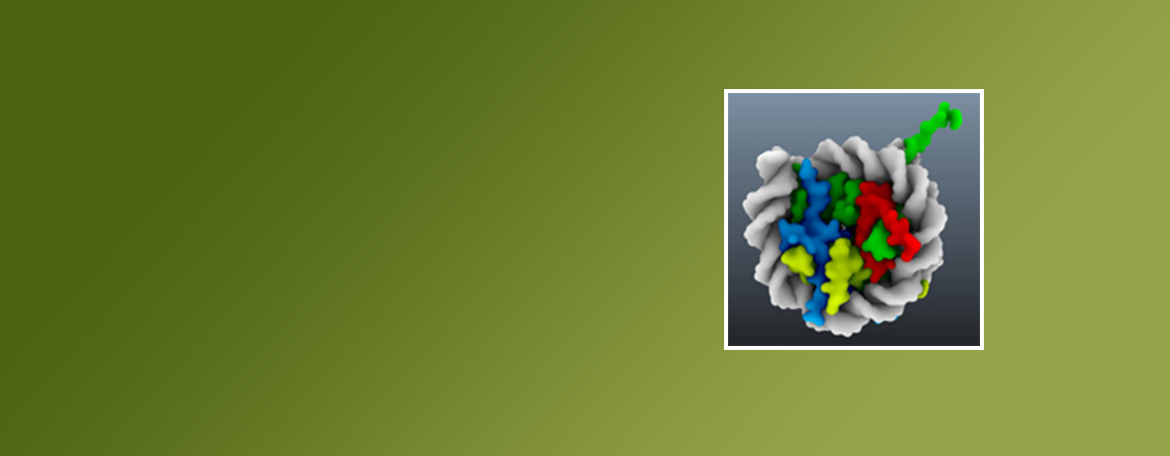
Bezsonova Lab
My expertise and research interests are in elucidating structure, dynamics and interactions of biological macromolecules using structural biology methods, including NMR spectroscopy and X-ray crystallography. The focus of my current research is on ubiquitin biology and the role ubiquitination plays in cancer development and gene silencing.

Korzhnev Lab
Our research focuses on protein assemblies involved in DNA damage tolerance pathways responsible for mutagenesis in eukaryotes, making use of cutting-edge structural biology methods to decipher their molecular mechanisms of action.

Hao Lab
The research of the Hao group aims to determine the physical and chemical principles underlying the structure/function relationships in biomedically important proteins, with a focus on ubiquitin E3 ligases that are altered in cancer and spore germination proteins that can cause food spoilage and food-borne disease as well as anthrax.
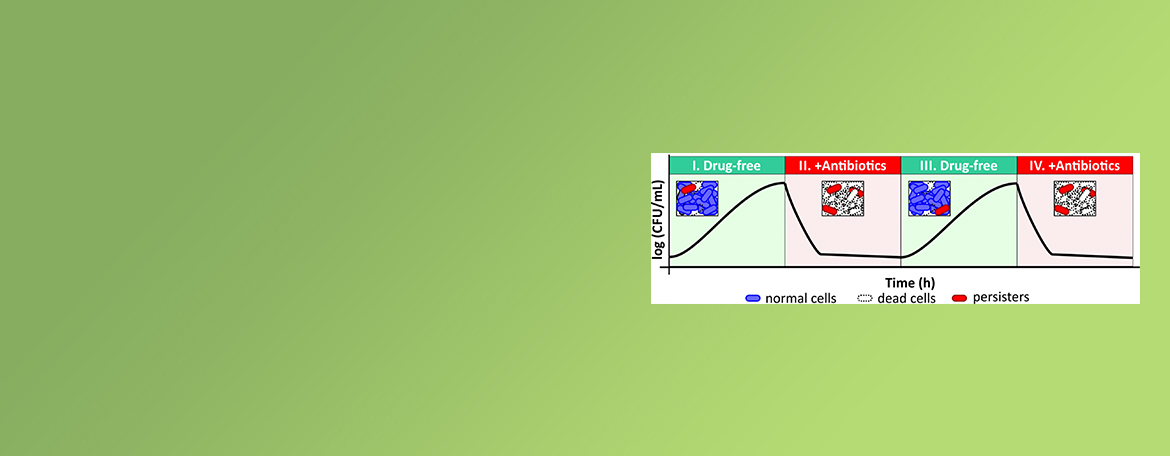
Mok Lab
Persisters are rare cell types in a bacterial population that can tolerate lethal doses of antibiotics that kill their genetically identical kin and can contribute to relapsing infections. The Mok lab is interested in investigating how bacteria respond to and survive treatment with antibiotics that target distinct cellular components under different growth niches.

Torti Lab
Our laboratory is interested in the relationship between iron metabolism and cancer. We examine the hypothesis that iron contributes to cancer using cell culture systems, animal models and patient databases, as well as through collaborative work with systems biologists.

Weller Lab
The Weller lab studies the mechanisms of HSV DNA replication, encapsidation and viral assembly and explores novel antiviral strategies aimed at inhibiting these processes. We are also interested in how HSV manipulates host stress responses such as DNA repair and the protein quality control machinery.

Caimano Lab
My research focuses on the pathogenic spirochetes Borrelia burgdorferi and Leptospira interrogans, the causative agents of Lyme disease and Leptospirosis, respectively. Our broad objective is to better understand the environmental signals, regulatory pathways and gene products required to maintain these unique bacteria within their enzootic and zoonotic life cycles and establish infection within the mammalian host.

Schuyler Lab
The Schuyler lab utilizes computational modeling to study binding and allosteric signalling in order to elucidate complicated disease mechanisms and inform drug design. Current work includes the study of SOD1 mutations that cause ALS.

King Lab
Cilia and flagella are microtubule-based organelles that have essential motile and sensory functions. The King lab studies the dynein molecular motors that are required to assemble and power these organelles using genetically and biochemically tractable model systems.

Radolf Lab
The Radolf laboratory focuses on the pathogenic spirochetes Treponema pallidum and Borrelia burgdorferi, the causative agents of syphilis and Lyme disease, respectively. The major objective of our syphilis research is to elucidate the ultrastructure and function of T. pallidum's highly unusual outer membrane and rare outer membrane proteins. With Lyme disease, our major objective is to elucidate the genetic programs that enable B. burgdorferi to transit back and forth from...

Ann Cowan
My work uses high temporal and spatial resolution fluorescence imaging methods to explore the dynamic behavior of molecules in living cells. Experimental results are integrated within the Virtual Cell mathematical modeling software environment to create three-dimensional computational models of a variety of different cellular events.
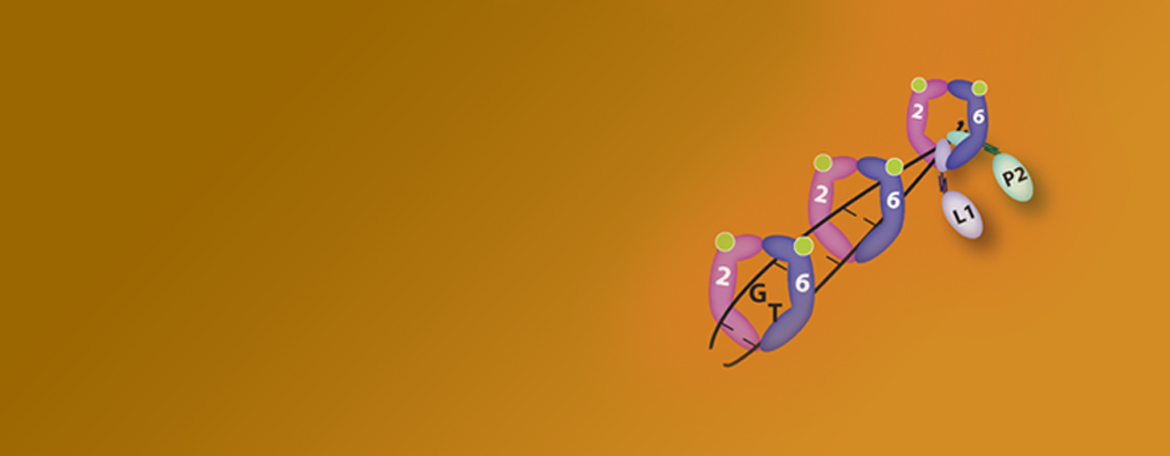
Heinen Lab
The Heinen laboratory is interested in the molecular mechanism of the human DNA mismatch repair pathway and in understanding why defects in this pathway are associated with the hereditary cancer predisposition disease Lynch syndrome.
Carson Lab
Our lab focuses on localization and translation of RNA molecules in ribonucleoprotein complexes called RNA granules, which are affected in neurological disorders such as fragile X tremor ataxia syndrome.
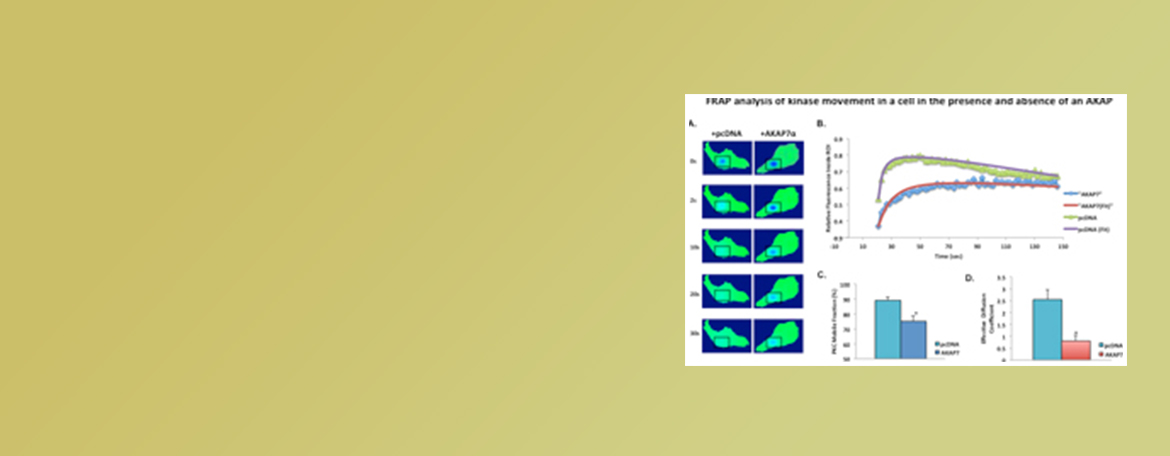
Dodge-Kafka Lab
We investigate the mechanisms utilized by scaffolding proteins to create specificity of phosphorylation. Our work encompasses many domains including enzyme kinetics, live cell imaging, cellular physiology, and computation modeling.

Gryk Lab
The Gryk lab focuses on solution NMR studies of biomolecular macromolecules, primarily proteins. A current emphasis is on software integration and data management for NMR data analyses - a project called CONNJUR.
 Research in the Molecular Biology and Biophysics department focuses on understanding the proteins and pathways affected in human disease. From the structural biology of cancer drug targets to the mechanistic interplay between viruses and human cells, MBB labs study the basic molecular processes underlying human illnesses.
Research in the Molecular Biology and Biophysics department focuses on understanding the proteins and pathways affected in human disease. From the structural biology of cancer drug targets to the mechanistic interplay between viruses and human cells, MBB labs study the basic molecular processes underlying human illnesses.
Our department can be divided into five thematic areas:
Spotlight
- Spotlight: August 2022Publications Gao, X., Swarge, B., Roseboom, W., Wang, Y., Dekker, H.L., Setlow, P., Brul, S. and Kramer, G. 2022. Changes in the spore proteome of Bacillus cereus in response to introduction of plasmids. Microorganisms. https://www.mdpi.com/2076-2607/10/9/1695 Hereditary hemochromatosis variant associations with incident non-liver malignancies: 11-year follow-up in UK Biobank. Atkins JL, Pilling LC, Torti SV, Torti […]
- Spotlight: July 2022On July 1, 2022, Dr. Steven Chou joined MBB as the newest tenure-track faculty member. Welcome Steve to MBB! His office is L3081, and his laboratory space is L3078. Stop by his office and get to know the new MBB space in the nicely renovated 3rd floor area. His laboratory is starting to look like a […]
- Congratulations on All the Fantastic New PublicationsBusy start to the summer. Publications Luxmi, R., Mains, R.E., Eipper, B.A., and King, S.M. (2022). Regulated processing and secretion of a peptide precursor in cilia. Proceedings of the National Academy of Sciences USA in press. Kristina N. Delgado, Jairo M. Montezuma-Rusca, Isabel C. Orbe, Melissa J. Caimano, Carson J. La Vake, Amit Luthra, Chris […]
Upcoming Events
-
Sep
9
Molecular Biology and Biophysics Seminar: Dr. Smita Gopinath 12:00pm
Molecular Biology and Biophysics Seminar: Dr. Smita Gopinath
Tuesday, September 9th, 2025
12:00 PM - 01:00 PM
Low Learning and Webex
Dr. Smita Gopinath, Assistant Professor, Immunology and Infectious Diseases, Harvard T.H. Chan School of Public Health
Title: TBA
Hosts: Dr. Wendy Mok and Dr. Sandra Weller
-
Sep
23
Molecular Biology and Biophysics Seminar: 12:00pm
Molecular Biology and Biophysics Seminar:
Tuesday, September 23rd, 2025
12:00 PM - 01:00 PM
Low Learning and Webex
-
Oct
7
Molecular Biology and Biophysics Seminar: 12:00pm
Molecular Biology and Biophysics Seminar:
Tuesday, October 7th, 2025
12:00 PM - 01:00 PM
ARB Large Conference Room and Webex
JC = Journal Club
WIP = Work in Progress talk
Quick Links
Contact Us
Department of Molecular Biology and Biophysics
UConn Health
263 Farmington Avenue
Farmington, CT 06030
Phone: 860-679-7682
Fax: 860-679-3408
Email: mbb@uchc.edu