CCAM's quantitative and interdisciplinary research addresses fundamental questions of how the spatial organization of molecules in cells is established and how it is utilized to control cell function. To answer these questions, faculty measure, model and manipulate the spatial organization, subcellular distribution, dynamic behavior, interactions and functions of specific molecules within the cell. Research areas include cell biology, biophysics, optical imaging, and cellular modeling.
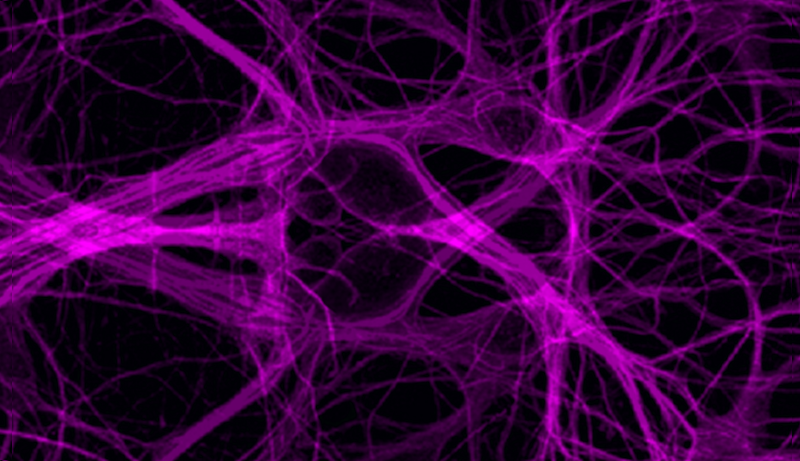
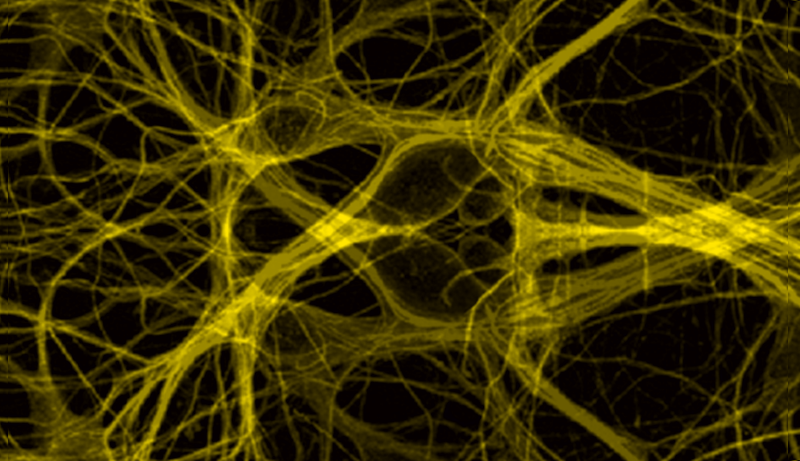
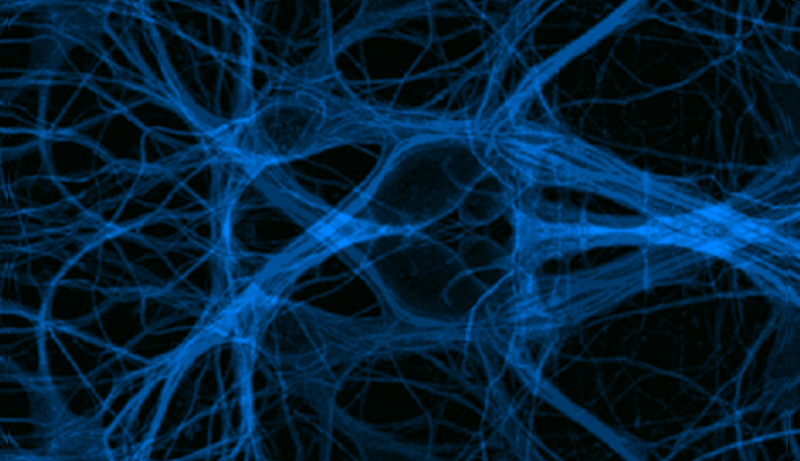
Cell Biology
Cytoskeletal Dynamics
Loew, Mayer, and Rodionov
Actin and microtubule dynamics play an important role in vesicular transport, e.g. melanophores in Xenopus melanocytes, and signaling transduction, nck signaling and macroscopic cellular behaviors such as cell motility, crawling and shape. Cellular, molecular, and biochemical approaches are used to understand the mechanisms of self-organization of cytoplasmic microtubules in living cells, regulation of microtubule dynamics, and cellular control over the activity of microtubule-based molecular motors. Biophysical modeling, such as solid and fluid mechanics, coupled with cellular imaging are used to probe the physical and biochemical mechanisms by which cells move, grow, and maintain their shape.
Cell Biology
RNA Trafficking
Carson
Research at CCAM has been at the forefront of utilizing advanced fluorescence imaging techniques to study RNA trafficking in cells. Research focuses on intracellular RNA trafficking in neural cells investigated through a combination of quantitative live cell imaging, fluorescence correlation spectroscopy, and mathematical modeling. Such studies have identified RNA granules- a RNA trafficking intermediate- and characterized spatial transriptional and translational regulation of RNAs localized to granules.
Cell Biology
Signal Transduction
Blinov and Mayer
Signal transduction pathways involve a complex series of molecular interactions in which cells transform events that occur at the cell surface into finely tuned signals for cell motility, division, or differentiation. Researchers at CCAM focus on detailed mechanistic modeling of EGFR signal transduction networks in single cells. A specific focus is the role of small adaptor proteins, Nck, in coupling signals from outside the cell to local rearrangements of the actin cytoskeleton, and identification of protein motifs integral to protein-protein interactions. Research in these areas stimulates advances in computational methods for studying signaling events and networks.
Biophysics
Biological Signaling Platforms
Loew and Mayer
Experimental manipulation of the location, size, and composition of signaling complexes is essential for building and validating quantitative models of spatially organized signaling networks and pathways. Researchers at CCAM are developing standardized methods to experimentally manipulate local concentrations and stoichiometries of membrane-bound and cytosolic signaling complexes (signaling platforms) in living cells. Experimentally manipulating the subcellular localization and/or local concentration of a factor in living cells provides insight to its function and resulting effects on biological activity. CCAM faculty use biological aggregrates, nano-fabrication and optical force to manipulate and study components of signaling networks. An example of a biological singaling platform is the creation of fusion transmembrane proteins that can be aggregated at the plasma membrane by monoclonal antibodies.
Cellular Modeling
Analysis and Simulation
Cowan, and Moraru
Complex imaging data contains a wealth of quantitative information, but it is often a non-trivial process to extract quantitative parameters from the data. Absolute or relative concentrations of a molecule within the cell, the fraction of molecules that are freely diffusing and their diffusion coefficients, kinetic constants for binding to large complexes, and velocities of directed motion are all parameters that can be obtained from imaging experiments. In most cases, cellular geometries preclude the use of simple analytic solutions to extract the relevant information. Spatial simulations within the same cellular geometry as the experiment provide a tool to determine such quantitative parameters. CCAM is building a suite of software tools specifically geared to these applications.
Cell Biology
Data Integration
Moraru, and Slepchenko
The development of quantitative models of biological processes often requires explicitly accounting for the experimental conditions involved in capturing experimental data. Thus experimental data is not only what is measured but what is used to take such measurements. In order to facilitate researchers' ability to rapidly create quantitative models of cellular physiology, faculty at CCAM are developing the computational technology ncessary to extract and reduce experimental data to standard formats that can be readily input into quantitative computational models. We develop software interfaces to data sources such as immunofluorescence images and Fluorscence Correlation Spectroscopy to derive spatial distributions, intracellular diffusion coefficients, concentrations and interactions. These data linkages require skills in computing and computation to accurately link experimental data directly to computational models.
Optical Imaging
Fluorescence Correlation Spectroscopy
Carson
Fluorescence Correlation Spectroscopy (FCS) can provide requisite quantitative data for analysis of molecular interaction networks underlying complex cell biological processes. (FCS) provides the technology to measure concentrations, diffusion coefficients, kinetic constants and conditional operators in living cells (Levin and Carson, 2003). This high resolution method of imaging the movement of fluorescent molecules in and out of a volume has been used to understand the complex regulation of RNA nuclear:cytoplasmic transport and RNA granule trafficking in neurons. In addition to the basic research in the areas of neuronal biology, RNA transport and trafficking, researchers are creating protocols that will make FCS easier for the scientific community to use.
Biophysics
Optical Probe Development
Loew
Fluorescent dyes are significant tools in the study of cellular processes. At CCAM novel chromophores from environmentally sensitive dyes are synthesized and characterized. Membrane associated styryl dyes are a particular interest in the study of transmembrane potentials and signaling events that occur due to voltage changes at the plasma membrane. Combining these dyes with additionally innovative methods of optics creates new quantitative ways of looking at cellular behaviors.
Biophysics
Single Molecule Imaging
Carson and Yu
Single molecule imaging allows cell biologists to analyze cellular behaviors at a new scale of resolution. Faculty at CCAM have used photo-switchable fluorescent protein for single-molecule tracking in live cells to analyze the dynamics of membrane-cytoskeletal interactions as well as properties of gene transcription and translation. Single molecule tracking involves the exploitation of chromophores for in vivo imaging, millisecond and nm resolution from 2-photon laser microscopy, deconvolution and tracking algorithms. Single molecule imaging, similar to FCS, is providing exceptionally fine resolution to cellular behaviors such that stochastic behaviors can be quantitatively measured.