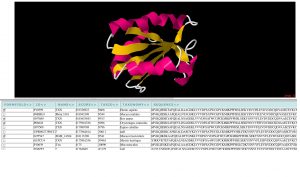
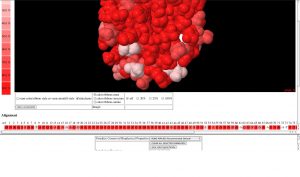
VENN is software tool for mapping protein sequence conservation among homologues onto a three dimensional structure of a protein (Vyas et al., 2009). VENN retrieves and interprets the PDB file to identify different structure chains. Protein sequences with sequence similarity to each chain are automatically retrieved from the EBI and a list of up to 500 such sequences is presented to the user. The user selects a subset of these sequences for alignment and subseqent display on the protein structure. VENN aligns the protein sequences, calculates a heatmap for conservation of each amino acid position and plots the heatmap onto the protein structure. Different strategies for selecting groups of sequences can be used to identify functional regions and specificity determinants in gene families.
VENN was originally developed by Jay Vyas using CONNJUR libraries; it is now hosted by the Schiller Lab.

Screenshots
References
Vyas J, Gryk MR, and Schiller. (2009). Venn, a tool for titrating sequence conservation onto protein structures. Nucleic Acids Res., 37, e124. PMCID: PMC2764419