Our research program focuses on the molecular and cellular processes that regulate the development of the cerebellum. The cerebellum is best known for its crucial rule in motor coordination. However, emerging evidence indicates that the cerebellum contributes to cognition and emotion as well. Accordingly, abnormalities in cerebellar development and function are linked to a wide variety of developmental brain diseases, such as autism spectrum disorder, schizophrenia, and language impairment. Despite the uniform appearance in its cytoarchitecture, the cerebellar cortex is subdivided into different functional areas resembling the cortical maps of the cerebrum. We are interested in the mechanisms that control the cellular heterogeneity and connectivity, which culminate in the parcellation of the cerebellar cortex. We use single-cell genomics and computational analysis to systematically study the gene regulatory networks that govern cell fate specification and cell differentiation in the developing cerebellum. Concurrently, we use mouse genetics to investigate how alterations of the regulatory network affect cerebellar development and function. Through these studies, our long-term goal is to gain insight into the causes of cerebellar-related diseases and develop therapeutic strategies to improve the quality of life of affected individuals.
Molecular Mechanisms of Cerebella Cell Fate Specification
The overall goal of this project is to investigate the regulatory logic underlying cell specification of various cerebellar cell types from neural progenitor cells. Our employ multi-pronged approaches as follows: 1) Use integrated single-cell transcriptomic and epigenetic analyses to infer developmental trajectories of cerebellar progenitors and to identify putative trans- and cis-elements that control cell state transition; 2) Reverse-engineer gene regulatory networks (GRNs) of each cerebellar cell type; 3) Computation GRN simulations to predict how perturbations of transcription factors impact cell fate decision; 4) Validate results of in silico studies with mouse genetic experiments.
Mechanisms of Assembly of Cerebellar Circuit
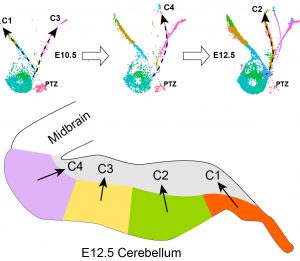
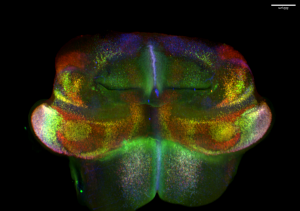
Abnormalities in cerebellar development, especially pathology and dysfunction of Purkinje cells, have been implicated in a wide variety of neurodevelopmental diseases, including ataxia, autism spectrum disorder, schizophrenia, and language impairment. Being one of the earliest-born cerebellar cell groups, Purkinje cells are believed instrumental in the development, function, and pathogenesis of the cerebellum. Evidence suggests the existence of Purkinje cell subtypes with distinct molecular features. However, the molecular mechanisms underlying the diversification of Purkinje cells remain poorly understood. Consequently, we lack an entry to assess the role of individual Purkinje cell subtypes. Through single-cell RNA and chromatin accessibility analyses, we uncovered at least nine molecularly distinct subtypes of Purkinje cells in the developing mouse cerebellum. These Purkinje cell subtypes contribute to different compartments in the developing cerebellum. Remarkably, the Purkinje cell subtypes display a characteristic combinatorial expression of Foxp1, Foxp2, and Foxp4, which belong to a subgroup of the forkhead-box transcription factor family. Mutations of human FOXP1 or FOXP2 are linked to speech disorders, autism spectrum disorder, and intellectual disability, indicating that these proteins coordinate the development of the neural circuits related to cognitive diseases. The specific goal of this project is to define the function of Foxp1/2/4 in the developing cerebellum by testing the hypothesis that Foxp1/2/4 coordinate the diversification of Purkinje cells, which in turn instruct the assembly of cerebellar circuit.