What are stem cells?
Why is it important to understand how stem cells are regulated?
- Understand how diseases occur. As stem cells keep producing tissue cells, dysregulation of them results in many disease conditions including cancers, tissue degeneration and aging. Better understanding of these conditions helps us to develop new therapeutic strategies for uncurable diseases.
- Generate healthy cells to replace damaged or lost cells. Stem cells can be used to produce specific cells that can be used in patients to regenerate or repair damaged tissues or organs. We need to generate cells in a culture dish by mimicking the environment in the real tissues. To do so, it is important to know how stem cells are regulated in the body.
What do we do?
We use Drosophila melanogaster (fruit flies) as a model system with a combination of genetics and various imaging techniques, including immunofluorescence, RNA and DNA FISH (fluorescent in situ hybridization), and long-term live-imaging. Owing to a simple anatomy and abundant genetic tools, this system is especially suited to discover previously unrecognized regulatory mechanisms.
Niche ligand diffusion ensures spatial control of the signal
In the Drosophila testis, germline stem cells (GSCs) are continuously producing sperm throughout the fly’s lifetime. When a GSC divides in niche, one daughter cell remains a GSC, while the other daughter cell starts differentiation. Niche derived BMP ligand has been thought to only activate GSCs but not other daughter cells despite of physical proximity of these cells. Using genetically-encoded nanobodies called Morphotraps, we blocked diffusion of BMP ligand without interfering with niche-stem cell signaling. Unexpectedly, we found that the diffusible fraction of ligand has opposite effect on the cells located inside and outside of the niche. It requires to promote differentiation of cells outside of the niche, while maintaining stemness of the cells inside of the niche, thereby ensuring spatial control of the niche with a single factor.
Current project is aiming to understand how the factor induces opposed responses in different cell types.
Related publication https://www.nature.com/articles/s41467-024-45408-7
Emerging function of somatic homolog pairing in stem cell regulation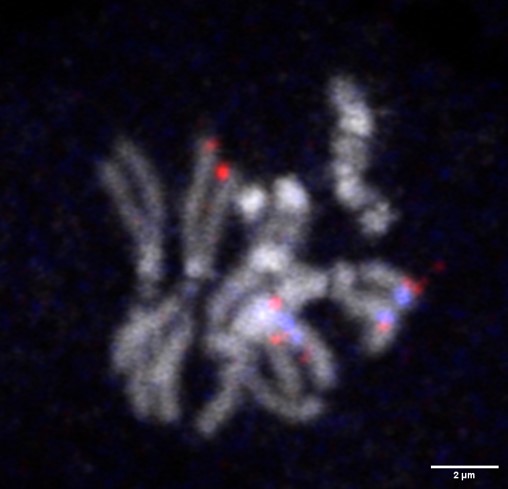
The pairing of homologous chromosomes is a fundamental process for meiotic recombination to exchange maternal and paternal genomic information. Homolog pairing also occurs in non-meiotic cells in a broad range of organisms including mammals. We recently found a change in the physical interaction of a homologous gene locus of a stem-cell specific protein, Signal transducer and activator of transcription 92E (Stat92E), during asymmetric division of GSCs. We found that the stat92E locus on two homologous chromosomes interacts closely (paired) in GSCs but becomes separated (unpaired) in the differentiating daughter cells. This "change" of pairing status seems to be required for prompt downregulation of this gene within next a couple of stages of differentiation. These results suggest a fascinating possibility that homolog pairing states is a potential mechanism that represents programmed gene expression changes which may occur in future. We are currently investigating more general roles of local homologous chromosome interaction on storage of cellar memory and cell fate determination.
Related publication https://www.nature.com/articles/s41467-022-31737-y