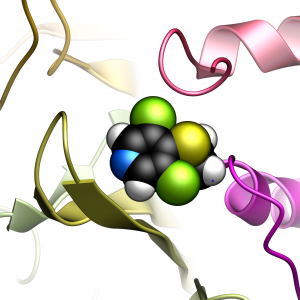
 USP7 Inhibition
USP7 Inhibition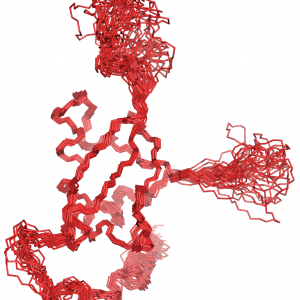 USP7 Structure and Dynamics
USP7 Structure and Dynamics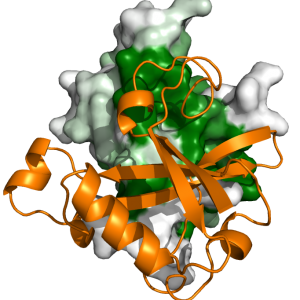 USP7 Molecular Interfaces
USP7 Molecular Interfaces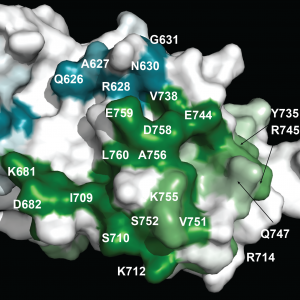 USP7 Substrate Recognition
USP7 Substrate Recognition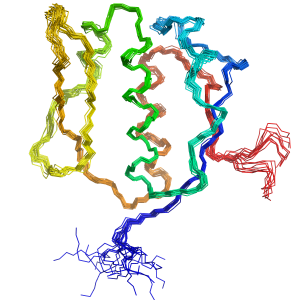 SLED Domain of Scml2
SLED Domain of Scml2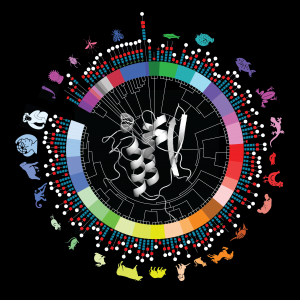 Evolution of SLED Domains
Evolution of SLED Domains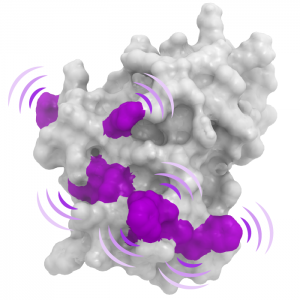 HIRAN Domain of HLTF
HIRAN Domain of HLTF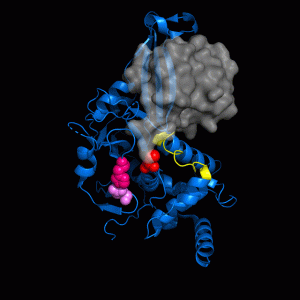 Conformational changes within the active site of USP7
Conformational changes within the active site of USP7
USP7 in cancer and neurodevelopmental disorders:
USP7 is a de-ubiquitinating enzyme responsible for the maintenance of several oncoproteins and tumor suppressors in the cell via the ubiquitin-proteasomal pathway. It is an attractive potential drug target for cancer therapy due to its role as a major regulator of p53 pathway. While upregulation of USP7 is linked to cancer, its loss causes a rare neurodevelopmental disorder - Hao-Fountain syndrome. We investigate molecular mechanisms of USP7 activation and substrate specificity to gain a detailed understanding of the USP7 enzymatic function and provide the structural basis for drug discovery.
Read more about our work:
-
DNA Polymerase ι Interacts with Both the TRAF-like and UBL1-2 Domains of USP7
-
USP7-Specific Inhibitors Target and Modify the Enzyme's Active Site via Distinct Chemical Mechanisms
- Structural Characterization of Interaction between Human Ubiquitin-specific Protease 7 and Immediate-Early Protein ICP0 of Herpes Simplex Virus-1
SUMO E3 ligase PML as a sensor of stress:
The promyelocytic leukemia protein (PML) is an E3 SUMO ligase involved in the regulation of various critical processes in the cell, including DNA repair, senescence, apoptosis, antiviral responses, and oxidative stress response. In response to oxidative stress and viral infections PML forms spherical compartments in the nucleus known as PML nuclear bodies (PML-NBs). Functionally, PML bodies may sequester, modify, or degrade partner proteins. Our laboratory studies the molecular mechanism of PML-NB formation in response to Herpes Simplex 1 (HSV1) infection. By characterizing the structure, dynamics, and oligomerization of PML protein we reveal PML features that make it a potent sensor of stress. This work may lead to the development of new effective HSV1 treatments and safer treatments for promyelocytic leukemia.
Read more about our research:
UBE3A in Angelman Syndrome:
Angelman Syndrome (AS) is a rare neurodevelopmental disorder caused by the loss UBE3A gene expression. In humans, UBE3A gene encodes three isoforms of the ubiquitin ligase E6AP. These isoforms differ only at their N-termini, with isoforms 2 and 3 having an additional 23 and 20 amino acid residues, respectively, that are not present in isoform 1. Data from the murine homolog, Ube3a, indicate differences in function and sub-cellular location among the three isoforms. However, the expression of all three protein isoforms in humans has only recently been demonstrated, and very little is currently known about human isoform-specific function and localization. We study structural and functional differences between the E6AP isoforms in humans.
Read more about our research:
Zn-binding AZUL domain of human ubiquitin protein ligase Ube3A