Murphy Lab
Our research focuses on endothelial cells lining the vasculature, aiming to better understand the critical role of these cells in mediating inflammatory and immune responses. Our work has revealed post-transcriptional regulation by alternative splicing in the response to inmate immune cell recruitment.
Splicing is increasingly recognized as an important contributor to human disease. Variations resulting from changes in splicing can produce proteins with entirely new functions. Although a number of critical endothelial proteins are alternatively spliced, little is known about the regulation and function of alternative splicing in the endothelium.
We are using genetic models to examine the impact of individual splicing changes on inflammatory responses within the vasculature, and bioinformatics and high-throughput genetic screens to identify and understand the role of RNA-binding splice factors regulating these splicing changes.
We work closely with a group of talented researchers in Vascular Biology, Immunology, Neuroscience and the Institute for Systems Genomics to achieve our objectives; this includes faculty within the Center for Vascular Biology and the Calhoun Cardiology Center and the collaborators in the Immuno-Cardiovascular Group.
This set of projects uses mouse models with endothelial-specific deletion of splice factors implicated by expression and motif analysis in the endothelial response to innate immune cell recruitment, and activity in regulation of endothelial inflammation in our in vitro CRISPR-KO screens. We hypothesize that these splice factors alter the endothelial transcriptome post-transcriptionally, through effects on RNA-splicing, stability or translation to modify the endothelial interaction with immune cells in chronic inflammation. Our recent publication, describing a key function for the splice factor Ptbp1 in endothelial NFkB signaling, myeloid recruitment and plaque formation is an example of this class of RNA-binding proteins. Several other proteins are being examined using these assays, and each appears to have unique effects on immune cell behavior.
Project 2:Define the signaling from innate immune cells leading to splice factor activation within the endothelium
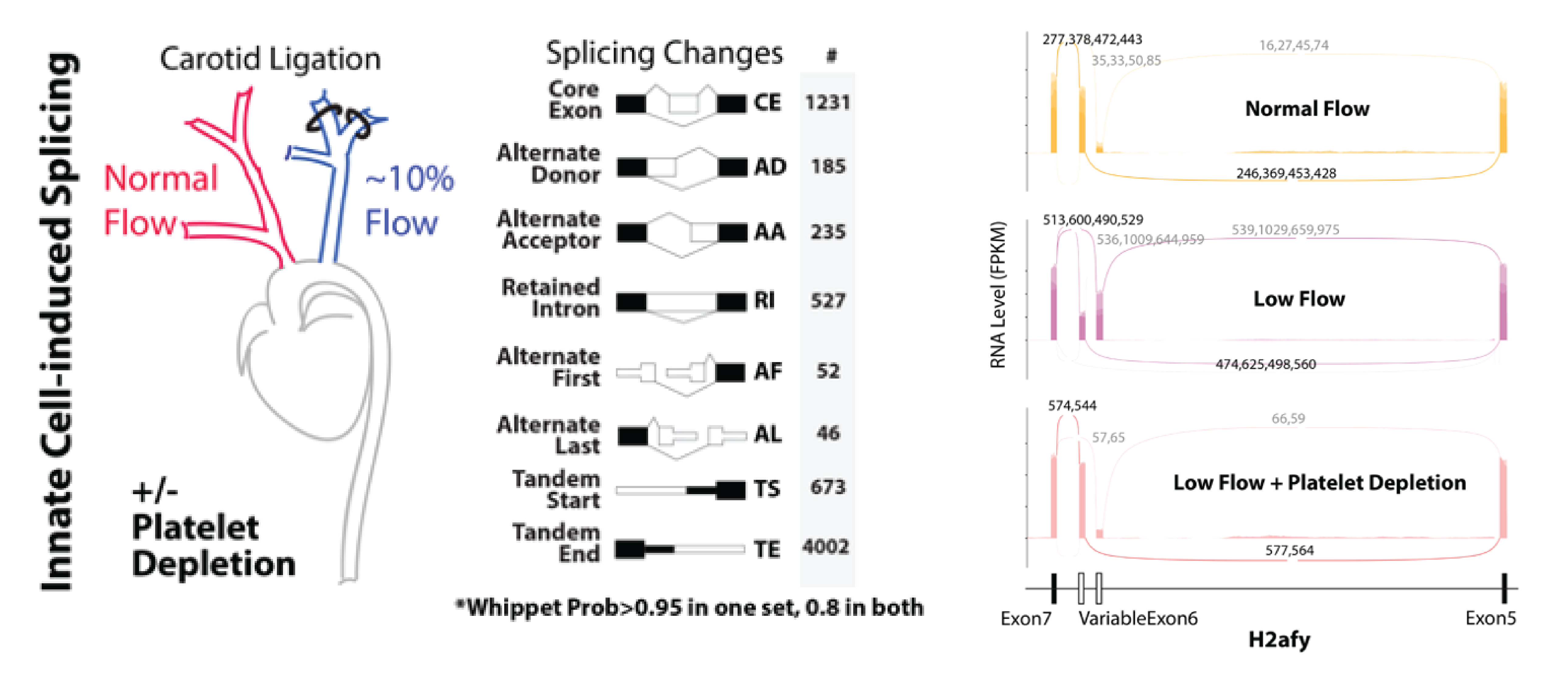
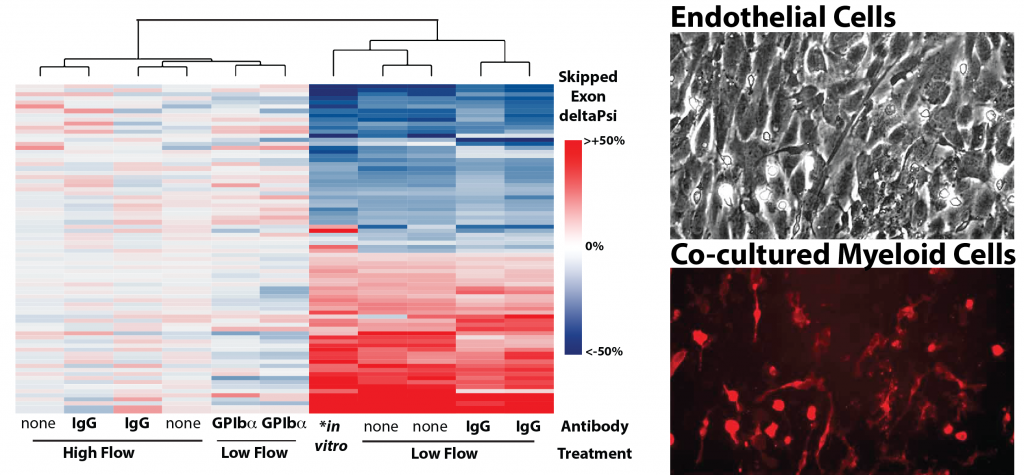
Figure shows heat map of splicing changes induced by innate immune cells in the arterial intima of the carotid artery (Murphy et al. eLife 2018). GPIba antibody was used to deplete platelets from mice prior to induction of low and disturbed flow. Note that, even in low flow conditions, platelet depletion blocks splicing changes in the endothelium. Adjacent images show, a DIC image of an aortic endothelial cell monolayer, which is overlaid with bone marrow derived monocytes from a reporter mouse (red).
Splice factor activity in the endothelium is induced by innate immune cell recruitment (platelet & myeloid dependent) in conditions of low and disturbed flow. We now know that activation of these splice factors is important in priming the endothelium for subsequent activation. We do not yet know the mediators of the signaling from innate immune cells to the endothelium which lead to increased expression and activation of these splice factors, but we do know that this process can be recapitulated in vitro, which provides us with an opportunity to identify these mediators. This project will focus on endothelial immune co-culture, screening for the effect of candidate mediator receptors on the endothelium for their effect on the expression and activation of the splice factors of interest.
Project 3: Identification of splicing changes induced by splice factor activation leading to effects on immune responses in vivo
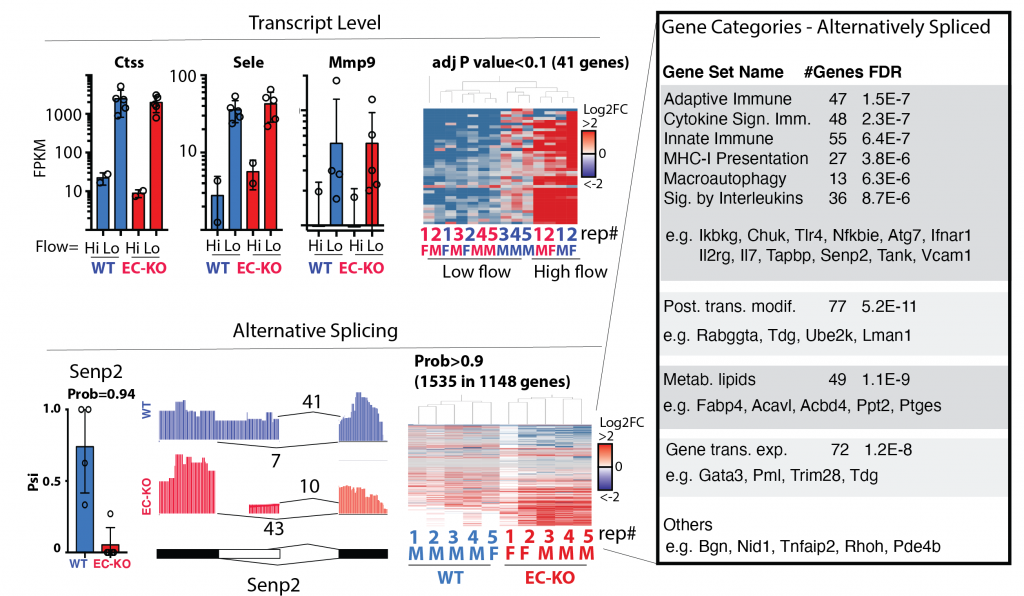
Figure shows the RNA-sequencing analysis from the carotid intima of mice exposed to disturbed flow or not. This splice factor had little effect on total RAN expression (upper panels, FPKM = Fragments Per Kilobase of transcript per Million mapped reads). However, there was a large effect on RNA splicing, primarily affected the genes and pathways in the inset.
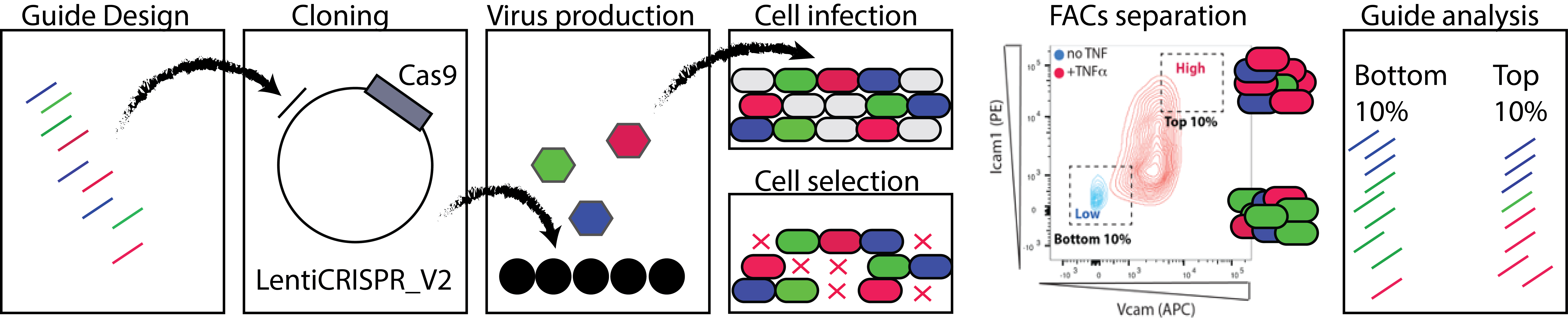
This set of projects is focused on analysis of altered splice isoforms within the endothelium induced by activation of the splice factors above. We have good data showing that, for some of these splice factors, there is little change in RNA expression when we look at all of the transcripts produced from a single gene, but that there are large changes in the specific splice isoforms that are produced. As these splice isoforms are enriched in pathways related to the immune phenotypes we have observed, we hypothesize that differential splicing of these transcripts contributes to the altered immune responses we have observed in vitro and in vivo. This work will use informatics and CRISPR-based screening approaches to define the key splicing events linking splice factor function to phenotype. Extrapolation of this work, to examine splicing QTLs (sQTLs) in human datasets will provide an important link to human disease.
Project 4: Interrogation of splice factor dysfunction in neurodegenerative disease using our novel methods of endothelial nuclei enrichment and single cell analysis
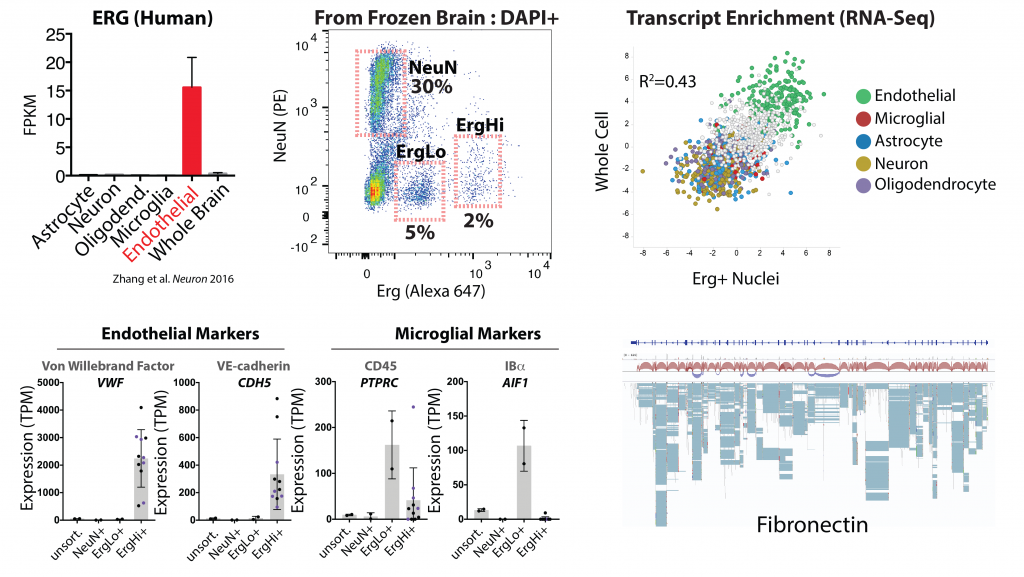
Figure cell-type specific expression of Erg in the brain in endothelial cells (from Zhang et al. 2016), and our use of an Erg specific antibody to isolate a high Erg+ population of endothelial nuclei from archived frozen brain tissues. RNA-sequencing analysis of these nuclei reveals enrichment endothelial specific transcripts (green) and a good concordance with whole brain endothelial cells. Lower panels show RNA expression levels of example markers of brain endothelial cells and microglia in the indicated sorted populations. The last panel shows analysis of the Fn transcript, and splice isoforms, in Erg+ nuclei. Details on these approaches can be found in our pre-print below.
Splicing dysregulation, caused by defects in RNA-binding splice factors (e.g. TDP-43 and MATR3), is considered a hallmark and potential driver of neuronal dysfunction and cognitive decline in Alzheimer’s (AD) and Related Dementia (ADRD). Although substantial evidence suggests that genetic causes of splicing dysfunction are not limited to neuronal cells, splicing defects in the endothelium have not been examined. As the endothelium is a principal component of the blood brain barrier, and defects in the blood brain barrier function increase with age and early in the progression of AD and ADRD, we believe that understanding splice factor function in the endothelium in aging and disease states could provide new approaches to targeting age and dementia related declines in cognitive function. We recently developed a method to specifically extract endothelial nuclei, based on the expression of an endothelial transcription factor in these nuclei (Erg), and are using this to profile the endothelial transcriptome across aging and disease states. This will be used to identify dysfunctional splicing patterns within the endothelium, and candidate regulatory splice factors. These will be functionally validated in vitro using our established screening assays and in vivo, using a novel combination of Br1-AAV for brain endothelial delivery of gRNA with a mouse model of endothelial specific Cas9 expression.
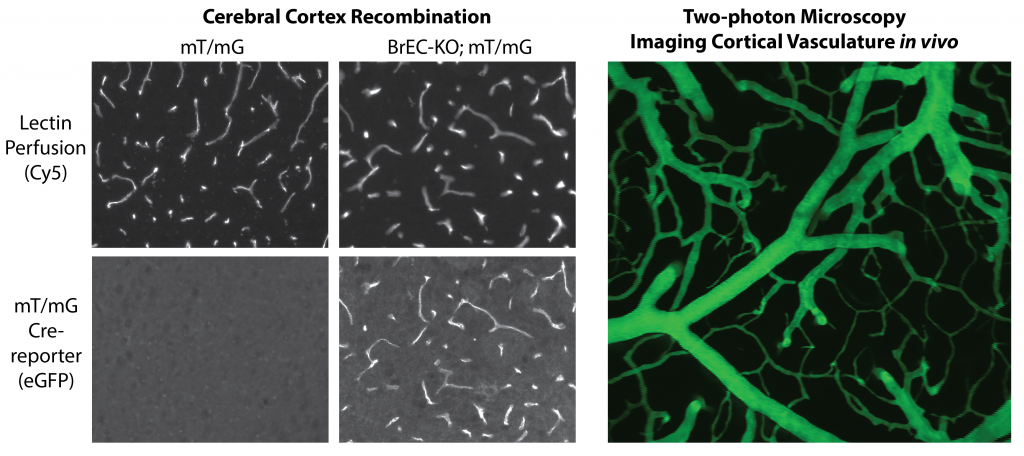
Project 5: Analysis of the impact of common disease-causing mutations on endothelial splicing patterns and function in mouse models
Figure shows Sclo1c1-CreER activity in brain endothelial cells, allowing the specific deletion of splice factors, such as Tardbp (TDP-43) in the brain endothelium for phenotypic characterization. Adjacent figure shows in vivo multiphoton imaging of dextran labeled vasculature through a cranial window. This approach is used to assess endothelial barrier functions, leukocyte recruitment and blood flow patterns in vivo.
Several genetic mutations are known to directly or indirectly affect TDP-43 (Tardbp) function during the progression of neurogenerative disease. These include mutations in TDP-43 itself, leading to increased cytoplasmic aggregation, as well as mutations in other pathways (e.g. Proganulin, GRN) which indirectly lead to TDP-43 dysfunction. Although earlier work in zebrafish morpholino knockdowns suggested a potential direct effect on vascular growth and development, very little is known about TDP-43 function within the brain endothelium. As several studies have indication that TDP-43 dysfunction in disease may not be limited to the endothelium, we propose that an analysis of endothelial cell functions will be important in understanding blood brain barrier dysfunction in neurodegenerative disease. We will use novel mouse models we have generated to examine TDP-43 functions within the endothelium and brain endothelium in vivo, and to examine the effect of commonly linked mutations, like those in GRN, to TDP-43 function in the endothelium. This work will focus on blood brain barrier properties, inflammatory status and responses, and neurovascular coupling. We will use a combination of mouse genetics, cellular and molecular characterization and intravital multi-photon imaging to achieve our goals.
You?
We are looking for talented researchers to join our team at all levels.
Steve Helming, M.S., Postbac Student 2023-2024, Current position : Postbac Student, Rosenberg Lab UConn Health
Sarah-Anne Nicholas, Ph.D., Graduate Student 2018-2023, Post-doc 2023 Current position : Scientist, Boehringer Ingelheim
Jordan Tyburski, B.A., Postbac Student 2021-2023. Current position : Medical student, UCONN
Sebastian Nagpal, B.A., Postbac Student 2020-2021. Current position : Actor, NY
Jessica Hensel, Ph.D., Graduate Student 2017-2021. Current position : Scientist, Vertex
Melissa Murphy, B.A., Postbac Student 2020-2021. Current position : Medical student, University of New England
Jordan Silva, B.A., Summer internship, 2019. Current position : Analyst, Health Advances
Brent Heineman, B.A., Postbac Student 2018-2019. Current position : Medical Student, UCONN
Xenia Bradley, M.D., Summer internship 2017.
- Amyotrophic lateral sclerosis and frontotemporal dementia mutation reduces endothelial TDP-43 and causes blood-brain barrier defects.
Cheemala A, Kimble AL, Burrage EN, Helming SB, Tyburski JD, Leclair NK, Omar OM, Zuberi AR, Murphy M, Jellison ER, Reese B, Hu X, Lutz CM, Yan R, Murphy PA. Sci Adv. 2025 Apr 18;11(16):eads0505. doi: 10.1126/sciadv.ads0505. Epub 2025 Apr 16. PMID: 40238886.
- Endothelial TDP-43 depletion disrupts core blood–brain barrier pathways in neurodegeneration.
Omar OMF, Kimble AL, Cheemala A, Tyburski JD, Pandey S, Wu Q, Reese B, Jellison ER, Hao B, Li Y, Yan R, Murphy PA. Nat Neurosci. 2025 Mar 14. doi: 10.1038/s41593-025-01914-5. Online ahead of print. PMID: 40087396.
- Endothelial Immunosuppression in Atherosclerosis : Translational Control by Elavl1/HuR.
Nicholas SA, Helming SR, Ménoret A, Pathoulas C, Xu MM, Hensel J, Kimble AL, Heineman B, Jellison ER, Reese B, Zhou B, Rodriguez-Oquendo A, Vella AT, Murphy PA. bioRxiv. 2023 Dec 14:2023.12.13.571184. doi: 10.1101/2023.12.13.571184. Preprint.. PMID: 38168388.
- Splice Factor Polypyrimidine tract-binding protein 1 (Ptbp1) is Required for Immune Priming of the Endothelium in Atherogenic Disturbed Flow Conditions.
Hensel JA, Nicholas SE, Kimble AL, Nagpal AS, Omar OMF, Tyburski JD, Jellison ER, Ménoret A, Ozawa M, Rodriguez-Oquendo A, Vella AT, Murphy PA. Proc Natl Acad Sci U S A. 2022 Jul 26;119(30):e2122227119. PMID: 35858420.
- A Method for Rapid Flow-cytometric Isolation of Endothelial Nuclei and RNA from Archived Frozen Brain Tissue.
Kimble AL, Silva J, Omar OM, Murphy M, Hensel JA, Nicholas SE, Jellison ER, Reese B, Murphy PA. Lab Invest. 2022 Feb;102(2):204-211. PMID: 34775494
- AtheroSpectrum Reveals Novel Macrophage Foam Cell Gene Signatures Associated With Atherosclerotic Cardiovascular Disease Risk.
Li C, Qu L, Matz AJ, Murphy PA, Liu Y, Manichaikul AW, Aguiar D, Rich SS, Herrington DM, Vu D, Johnson WC, Rotter JI, Post WS, Vella AT, Rodriguez-Oquendo A, Zhou B. Lab Invest. 2022 Feb;102(2):204-211. PMID: 34775494
- Identification of Splice Regulators of Fibronectin-EIIIA and EIIIB by Direct Measurement of Exon Usage in a Flow-Cytometry Based CRISPR Screen.
Hensel J, Heineman B, Kimble A, Jellison A, Reese B, Murphy PA. Sci Rep. 2021 Oct 6;11(1):19835. PMID: 34615942
- Alternative Splicing of FN (Fibronectin) Regulates the Composition of the Arterial Wall Under Low Flow.
Murphy, PA .; Murphy PA, Jailkhani N, Nicholas SA, Del Rosario A, Balsbaugh J, Begum S, Kimble A, Hynes RO Arterioscler Thromb Vasc Biol. 2021 41(1):e18-32. PMID: 33207933
- Direct CD137 costimulation of CD8 T cells promotes retention and innate-like function within nascent atherogenic foci.
Xu MM, Ménoret A, Nicholas SE, Gunther S, Sundberg EJ, Zhou B, Rodriguez A, Murphy PA*, Vella AT*. Am J Physiol Heart Circ Physiol. 2019 April. PMID: 30978132
- Activated T effector seeds: Cultivating atherosclerotic plaque through alternative activation.
Xu MM, Murphy PA, Vella AT. Am J Physiol Heart Circ Physiol. 2019 Mar 29. doi: 10.1152/ajpheart.00148.2019. [Epub ahead of print] PubMed PMID: 30925075.
- Endothelial FN (Fibronectin) Deposition by α5β1 Integrins Drives Atherogenic Inflammation.
Al-Yafeai Z, Yurdagul A Jr, Peretik JM, Alfaidi M, Murphy PA, Orr AW.
Arterioscler Thromb Vasc Biol. 2018 Nov;38(11):2601-2614. doi: 10.1161/ATVBAHA.118.311705. PubMed PMID: 30354234; PubMed Central PMCID:
PMC6209122. - Alternative RNA splicing in the endothelium mediated in part by Rbfox2 regulates the arterial response to low flow.
Murphy, Patrick A.; Butty, Vincent L.; Boutz, Paul L., Begum, Shahinoor; Kimble, Amy L.; Sharp, Phillip A; Burge, Christopher B.; Hynes, Richard O. eLife 2018 Jan 2;7. pii: e29494.
- Integrin-targeted cancer immunotherapy elicits protective adaptive immune responses.
Kwan, Byron H.; Zhu, Eric F. ; Tzeng, Alice; Sugito, Harun R.; Eltahir, Ahmed A.; Ma, Botong; Delaney, Mary K.; Murphy, Patrick A.; Kauke, Monique J.; Angelini, Alessandro; Momin, Noor; Mehta, Naveen K.; Maragh, Alecia M.; Hynes, Richard O.; Dranoff, Glenn; Cochran, Jennifer R.; Wittrup, K. Dane Journal of Experimental Medicine 2017 Jun 5;214(6):1679-1690
- Mouse Models of Cerebral Arteriovenous Malformation.
Nielsen, Corinne M; Huang, Lawrence; Murphy, Patrick A; Lawton, Michael T; Wang, Rong A Stroke 2016 Jan;47(1):293-300
- Tumor angiogenesis in the absence of fibronectin or its cognate integrin receptors.
Murphy, Patrick A; Begum, Shahinoor; Hynes, Richard O PloS one 2015 Jan;10(3):e0120872
- Constitutively active Notch4 receptor elicits brain arteriovenous malformations through enlargement of capillary-like vessels.
Murphy, Patrick A; Kim, Tyson N; Huang, Lawrence; Nielsen, Corinne M; Lawton, Michael T; Adams, Ralf H; Schaffer, Chris B; Wang, Rong A Proceedings of the National Academy of Sciences of the United States of America 2014 Dec;111(50):18007-12
- Alternative splicing of endothelial fibronectin is induced by disturbed hemodynamics and protects against hemorrhage of the vessel wall.
Murphy, Patrick A; Hynes, Richard O Arteriosclerosis, thrombosis, and vascular biology 2014 Sep;34(9):2042-50
- Notch4 normalization reduces blood vessel size in arteriovenous malformations.
Murphy, Patrick A; Kim, Tyson N; Lu, Gloria; Bollen, Andrew W; Schaffer, Chris B; Wang, Rong A Science translational medicine 2012 Jan;4(117):117ra8
- Endothelial Notch signaling is upregulated in human brain arteriovenous malformations and a mouse model of the disease.
Murphy, Patrick A; Lu, Gloria; Shiah, Steven; Bollen, Andrew W; Wang, Rong A Laboratory investigation 2009 Sep;89(9):971-82
- Endothelial Notch4 signaling induces hallmarks of brain arteriovenous malformations in mice.
Murphy, Patrick A; Lam, Michael T Y; Wu, Xiaoqing; Kim, Tyson N; Vartanian, Shant M; Bollen, Andrew W; Carlson, Timothy R; Wang, Rong A Proceedings of the National Academy of Sciences of the United States of America 2008 Aug;105(31):10901-6
- Complete list in PubMed
September 2016 - the Murphy lab is up and running!
July 2018 - Congratulations Jess on passing your qualifying exam with flying colors.
October 2018 - Thanks to the North American Vascular Biology Organization, for recognizing Sarah-Anne's work with a Travel Award to the 2018 meeting. It was a great meeting!
October 2018 - Nice work Jess, on winning a poster prize at the 2018 North American Vascular Biology Organization meeting in Newport!
April 2019 - Congratulations Maria Xu, on your graduation from the Ph.D. phase of your program. I look forward to hearing about your next accomplishments, it was a pleasure working with you and Dr. Vella to uncover new ways in which activated T cells are retained and re-activated within atherosclerotic plaque.
May 2019 - Thank you, to the American Heart Association, for the 2019 Innovative Project Award to apply our screening approach to identify endothelial RNA-binding proteins involved in the regulation of inflammatory responses.
June 2019 - Congratulations Sarah-Anne on passing your qualifying exam, I'm looking forward to your outstanding PhD work!
September 2019 - Happy to share my thoughts on the post-doc to faculty transition through the North American Vascular Biology Organization "Lessons Learned"; NAVBO has been a key component of my success in research and I am happy to have a chance to give back!
September 2019 - We appreciate the recognition by the North American Vascular Biology Organization as "Lab of the Month".
Dec 2019 - Thank you, to the American Heart Association, for funding Sarah-Anne's graduate work on endothelial Elavl1 functions in adaptive immune responses.
Dec 2019 - Thank you, to the American Heart Association, for funding Jess's graduate work on the regulation of NFkB responses by RNA-binding proteins.
May 2020 - We were honored to be considered a finalist for the Irvine H. Page Junior Faculty Research Award from the American Heart Association, and congratulations to Hanrui Zhang of Colombia on winning the award.
May 2020 - Thank you, American Heart Association, for the Paul Dudley White International Scholar Award for the top ranked submitted abstract from a United States lab to the 2020 Vascular Discovery Meeting.
August 2020 - We are grateful for RF1 funding from the NIH's National Institute of Neurological Disorders and Stroke, to support our work on splice factor function in the endothelial cells of the blood brain barrier in Alzheimer's and related diseases. We are incredibly enthusiastic about pushing boundaries over the next five years in work with our collaborators Riqiang Yan (UCONN Health), Li Gan (Weill Cornell) and Chris Schaffer (Cornell). Watch this space!
September 2020 - excited to present three different projects at the North American Vascular Biology conference, and to hear about other exciting work on immune cell interactions with the endothelium, and endothelial contributions to atherosclerosis and neurodegenerative disease.
October 2020 - congratulations Sarah-Anne, on a great talk at the 2020 NAVBO meeting, and congratulations Jess on a great poster presentation, and a poster award!
January 2021 - we welcome Ashok as a postdoc to the lab!
February 2021 - We are grateful for R01 funding from the NIH's National Heart, Lung, and Blood Institute to support our work to examine set of RNA-binding proteins linking to transposon-derived sequence in endothelial inflammatory responses. We hope to shed light on this important group of RNA-binding proteins and their functions in physiology and disease.
May 2021 - congratulations Jess, on your graduation and new position at Vertex.
September 2021 - welcome Omar and Olivia, as new graduate students in the lab!
October 2021 - had a great time at the 2021 North American Vascular Biology Organization meeting and round-table discussion on Cell-Matrix Interactions in Vascular Remodeling with Wayne Orr, Martin Schwartz, Sophie Astrof, Brenda Lilly, Jesus Torres-Vazquez and my Chair, Linda Shapiro, and chairing exciting nano-talks session with Joyce Bischoff
July 2022 - welcome Chris, as a new graduate student in the lab!
July 2022 - had a great time at our first Gordon post-transcriptional regulation conference, and presenting work on Ptbp1 in the endothelium - will be back again!
August 2022 - Thank you, to the NIH NHGRI (and Charles Lee and Brenton Graveley, for funding Chris's graduate work on endothelial RNA-binding protein involvement in graft vs host injury!
October 2022 - The lab had a great time traveling to Cambridge to celebrate Richard Hynes Lasker award, and dinner in town
December 2022 - Thank you, to the American Heart Association, for funding Omar's graduate work on the dose dependent effects of TDP-43 loss on the brain endothelium in neurodegeneration.
March 2023 - had a great time at our first Cold Spring Harbor Brain Barriers conference - and congratulations to Ashok on his poster prize, and Omar on a great presentation!
May 2023 - welcome Stephen as a post-bac and MS student to the lab.
June 2023 - Wonderful celebration of Richard Hynes and the seminal discovery of LETS (large external transformation sensitive protein), now known as Fibronectin 50 years ago.
July 2023 - had a great time at our first Cold Spring Harbor post-transcriptional regulation conference - will be back again!
July 2023 - welcome Swati as a post-bac student to the lab, and great lab outing for drinks and yard games.
September 2023 - congratulations Sarah-Anne, on defending your thesis and your PhD - we look forward to hosting your postdoc for the completion of your work!
September 2023 - We are grateful for RF1 funding from the NIH's National Institute of Neurological Disorders and Stroke to support our work with Martin Schwartz to examine the regulation of FLT-1 splicing and contributions to Vascular contributions to cognitive impairment & dementia (VCID).
October 2023 - so much fun bringing the lab the North American Vascular Biology conference in Newport RI, and excellent oral presentations by Sarah-Anne and Omar, and poster presentation by Ashok
October 2023 - welcome Sam, as a new graduate student in the lab!
June 2024 - Congratulations Sam on passing your qualifying exam, and being award the Esperance Fellowship in Personalized Nutrition. We appreciate the support of Roger Newton ’74 MS, ’05 (HON), and the Nutritional Science Program at UConn Storrs for their support!
June 2024 - Ashok and Omar enjoyed the Gordon Brain Barriers conference in New London, congratulations on Ashok on your talk, travel award, and poster award, and to Omar on an outstanding presentation. Thank you to the Brain Barriers meeting for your support of our work!


July 2024 - Omar, Chris and Patrick enjoyed the Gordon post-transcriptional conference in Portland, an awesome meeting and great place to get new ideas!
August 2024 - The lab had another fun outing to the Talcott Collective, a great place for frisbee golf and some refreshments!

October 2024 - so excited for Patrick's first invited talk at the North American Vascular Biology Meeting at Asilomar in Monterey, CA. This is particularly meaningful, as Patrick has attended this meeting since graduate work with Rong Wang at UCSF in 2004. Thank you Dr. Alex Adam and Dr. Gabby Fredman for the invitation. And congratulations Omar his talk and travel award, and Ashok on his excellent presentation


October 2024 - a treat to be back on the UCSF campus to give a talk and meet with Rong Wang.

November 2024 - congratulations Omar on your accepted paper to Nature Neuroscience.
January 2025 - so nice to see Ashok representing the lab internationally, through his trip to India and talks at Indian Institute of Science Education and Research (IISER) Bhopal, Pondicherry University and Bengaluru City University. See a TV interview from his visit here.
February 2025 - congratulations Ashok, on the acceptance of your paper at Science Advances!
March 2025 - congratulations Omar, on your successful defense!
April 2025 - congratulations Ashok, on your talk, and Emily on your poster presentation. Thanks to the CSH Brain Barriers group, it was a fun meeting!